DeepGANnel …finally published …and here is a blog about it. So Blogging 101 says “your blog post should have a point”. So this was my first stumbling point. …what would be my point? It could, of course, be the power of AI (that’s what we use in the paper), or the genius of our team that developed the idea? Well, OK taking all that as read; how about a blog about the “Trials and Tribulations of Publishing a Crossover paper?” It will be preaching to the converted I expect. Long ago, as an early career scientist (PDRA), I submitted a nice little pharmacology paper to Journal of Physiology. Senior author shall remain nameless. This paper was rejected and this was somewhat of a shock to the said Senior Author: “I have never had a Journal of Physiology paper rejected before” he pronounced. A then colleague (now wife) whispered… “well he has now” which gave us all a good titter. The fail, was probably, not just my fault to be fair, but the aura of an impending change of tide. I bet the days are gone where a few influential Senior scientists have their pet journals with guaranteed publication. But here is the story of DeepGANnel's publication which as been... quite a journey!
I figured after chatting with Yalin, Jaume and Frans, oooh around 2018(?) that a deep neural network would probably be pretty good at identifying single molecule level activity in ion channel data records. Thing is, by eye, you can often see the openings and closings of these proteisn, and sort of filter out the crap in your head. But to have an algorithm do this is pretty tricky…. So I presumed some AI might be just the ticket to do this. It works in our hands, yes, we’ve had deployment problems, changes to the underlying TensorFlow package we used etc…. But the basic principle is sound. Published here. Blogged previously here.
But the thing that makes this development really tricky is the need for masses of labelled training data. Not just for ion channels, but scFRET, Nanopore or ECG etc. You can’t just record these training data on a regular rig and go, because you don’t have the “ground truth” necessary for supervised training… essentially, you have no way to know whether a molecule was really open or closed to check if the AI actually worked. We found some devious methods to get around that, but it left Numan and I wondering whether you could actually use a different neural network to create the training set with which you could later train a detection network?! …so a simple idea was to use a GAN (first heard of after Numan/Jaume told me about them I think).
Now the clever bit is really not our usage of a GAN, but the very idea of a GAN. Here’s how they work. One neural network draws essentially a random picture of a thing…. A second neural network tries to see if it looks like the real thing or not. They then wrangle against each other until the picture gets more and more realistic. Forger versus Art Inspector...
So to deploy for electrophysiological data; we kind of turn a graph into a picture and then off you go. Turning a graph into a digital picture and visa versa is really trivial since of course, in computing terms; a picture is just a matrix of numbers anyway.
One of the fun (relatively!) things about these networks is you can watch the neural networks “learn” in real time. I’ve made a couple of cartoons of the training process here: First the generator (forger) spews out nonsense…. But the discriminator (inspector/or art critic) keeps correcting it and gradually the generator starts to learn to produce the desired output.
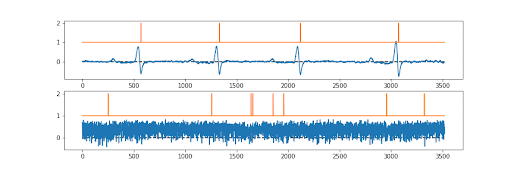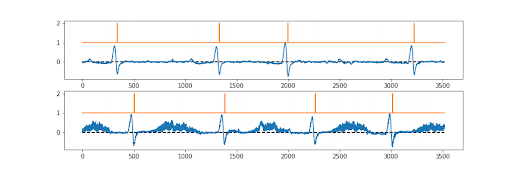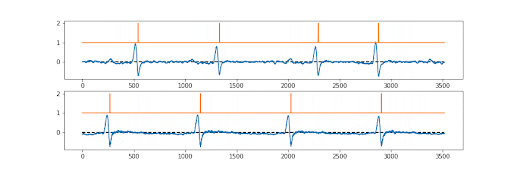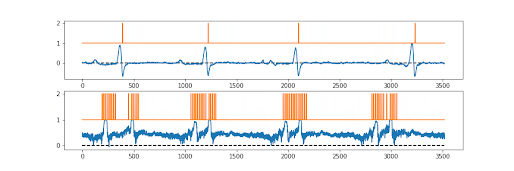
The first draft models were created quickly by Numan, but there was a problem. We needed both the ion channel signal… and the underlying ground truth to be perfectly synchronised. There was, so far as we (or the reviewers!!) were aware, no existing way of doing this? Sure various methods for associating the picture (say 512x512) produced with a particular label (cat vs dog vs car), but not a pixel by pixel ground truth. So the “simple” trick we used was essentially to make a picture that was 512x2 pixels in shape. Sort of a two lane picture. The first 512 pixel lane is the current converted to a pixel grey scale value…. The second 512 pixel lane is the ground truth; probability of a channel being open or closed, again converted to pixel greyscale. So they train perfectly together.
And it works! We also tested it with an ECG record, as you can see above, it was fine with this too. The signal is not absolutely perfect, but pretty good. Anyway; now just needed to publish it.
Time Table:
Beginning of the year, 2019… tossing around these ideas for synthesis of electrophysiological data with deep networks (Jaume, Numan and I). Firstly using, so called “autoencoders” which rand quite nicely with 1 channel ECG data. That was February 2019. Then we started to try with the GAN approach.
13th December 2019 the name of the network; “Deep-GANnel” is coined…. Feb 2020 Numan published a conference abstract on this, but still just generating one channel of (unlabelled) data. The idea at this point was to then feed the output into a new network that would label it, datapoint by datapoint. For me this didn’t really cut the mustard; what we really needed was this full annotation coming right out of the generator. So we treated, as described above, the image then as a 512x2 (in fact many different widths) and hey presto it worked!
Initially, we thought we just wanted it to generate any old ion channel data, so rather foolishly, we fed a wide range of different ion channel phenotypes in. This was actually an epic fail. It was like feeding in the images of a horse and a monkey and expecting it to simulate a realistic animal….. no you want it to produce horses right? ….so just feed it horses. We realised you needed to feed in a seed of the exact type of ion channel you wanted it to simulate. Now it really worked well.
17th April 2020. First draft of the paper ….
25th June 2020. Submission numero uno: Frontiers in SomethingorOther.
10th July 2020
They’d had it for over a month; could not even find an appropriate editor and reviewer set. AI-versant ion channel physiologists obviously far too rare!
10th August 2020…. Frontiers (lovely office staff) still not found an Editor comfortable with dealing with this!! …So I officially withdrew Frontiers Submission to go elsewhere.
14th August 2020…. submitted to PlosONE!!
2nd Sept 2020… PlosONE start to admin process DeepGANnel.
2nd October 2020…. Rejected!!!! …moderate rejection. To me there are three possible responses to paper submission. 1) Never darken our door again, 2) Do a load more stuff… 3) Make some little changes. Option 4) “Sure!!”. Is not something I have ever seen!!!! So of course, there were some silly comments in the rejection (along with very good points!!), there always are, I don’t take it personally. I may have been unclear, they may have misunderstood, I may have misunderstood them etc etc…. But the main thing they wanted was statistical evidence our methods were better than previous and wider examples of use (we only gave one previously). Of course the first of these is strictly impossible. There is no statistical measure of “better” and so we added a whole series more ion channels, and also added UMAP and TSNE clustering to show, more objectively, how realistic the data are. Also, and this is something I enjoyed including… a table of strengths and weaknesses of our method. It was actually really nice to just ‘fess up to the limitations”!!! Less pressure. So often in publication, you are trying to make something look as good as possible. Total openness is refreshing!! All the data; all the code, are on GitHub and FigShare. Despite the disappointment the original rejection brought…. The next version is oh so much better!!!! So thanks to the co-authors AND the editor/reviewers for this …and I learned some new techniques in those fancy clustering methods. As always, I was introduced to them by Jaume.
Took over a year, all the extra channels and experiments, to go again.
9th December 2021
Third Submission:
OK, Admin fail here…. Some error in the format of our resubmission. I think I highlighted changes, but tracked changes were necessary.
12th December 2021
Submitted again, with the change in format. This time makes it through the office checks by 14th December 2021.
14th January 2022
Original Editor no longer able to be involved. For all I know, they might not be well or just too busy. Hope the latter, but never will find out.
21st January 2022
A new editor found!! Thanks so much!!
21st February 2022… REJECTED!. but a soft rejection really, kind of class “3” from above.
25th February 2022… resubmitted….
7th April 2022… admin office checks completed in PlosONE office and can move to editor… OMG. This means 6 weeks after submitting, we were told that it had passed formatting checks and would now be assigned to an editor, who would choose reviewers…. My heart sunk: Yet another delay!! This usually takes a couple of days max!
But just 4 days after it landed back on the Editor’s desk…. Yay… deep joy!
11th April 2022 ACCEPTED… notwithstanding technical requirements, releasing the data we promised we would, clarifying funding etc. a further month of fiddling about with that type of thing. And proofs, as much as you get with PlosONE.
10th May 2022….. fully published…. So it was, off and on about 3 years. Actually, by my standards this is typical or rapid :-)
And now on to the next paper…..